The identification of glycosphingolipid structures in a biological sample is a non-trivial task requiring special expertise and experience. Established strategies rely on the release of the glycan from the lipid and the identification of both portions separately. However, in this approach the information which glycan is attached to which lipid is lost.
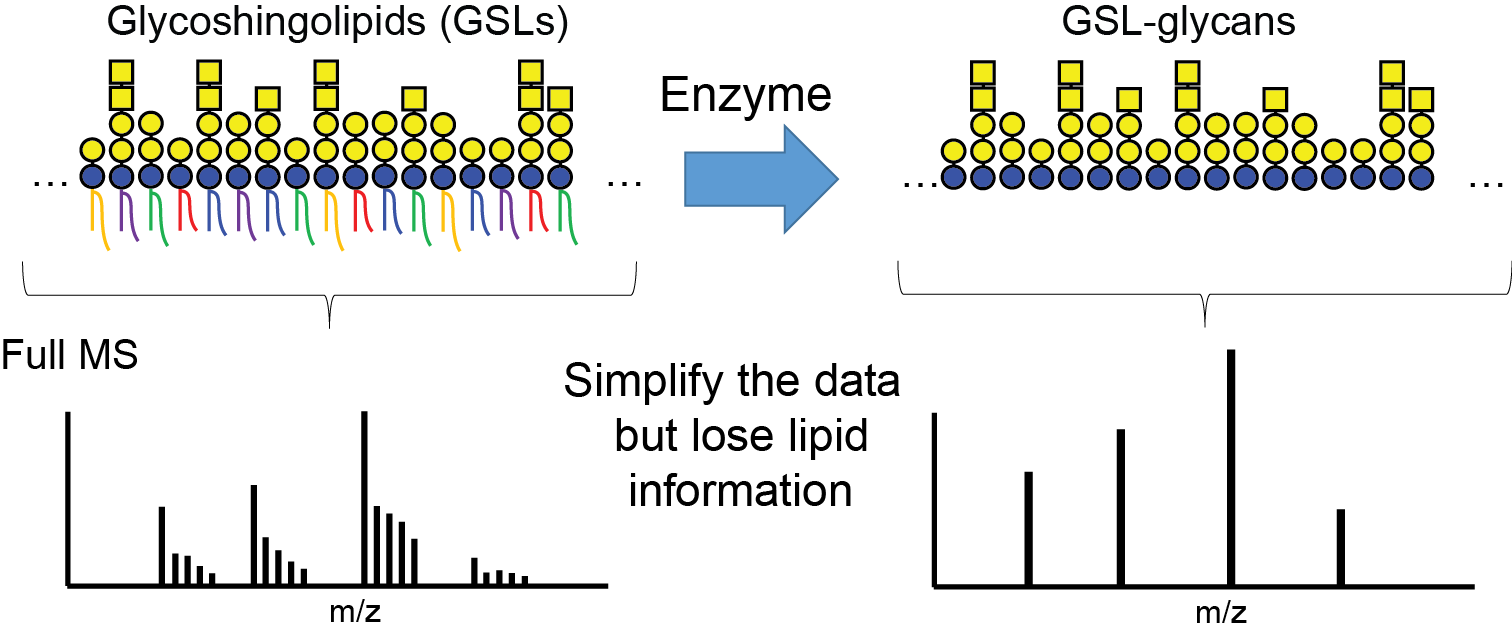
Therefore, we have developed an improved analytical method to profile glycosphingolipids (GSLs) that allows us to efficiently recover GSLs from a variety of biological materials.
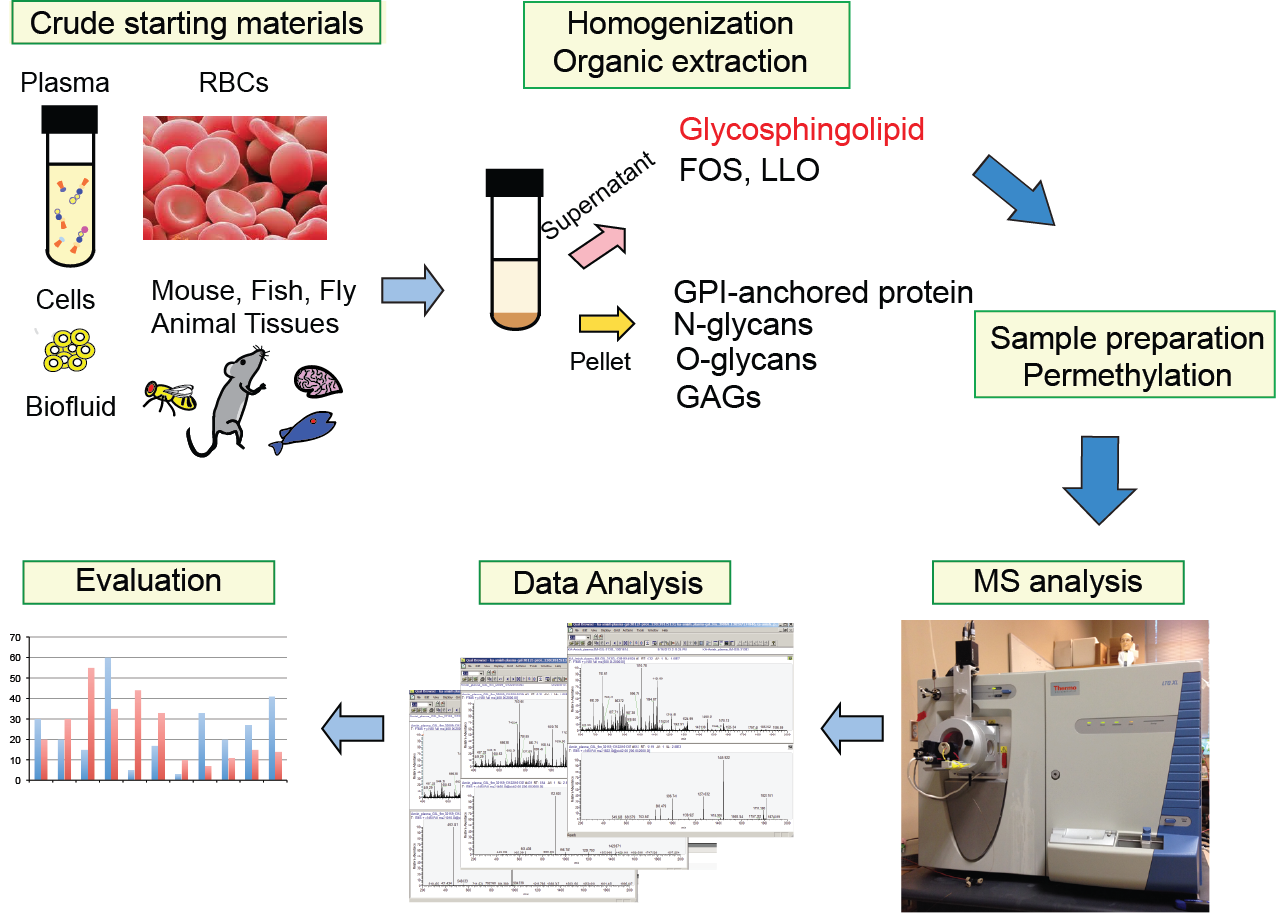
Our method yields miltiple glycomes from a single aliquot of starting material. GSLs will be analyzed by mass spectrometry following permethylation. Our MSn workflow allows us to characterize both glycan sequence and the composition of lipid (ceramide) moiety comprising of sphingosine and fatty acid.
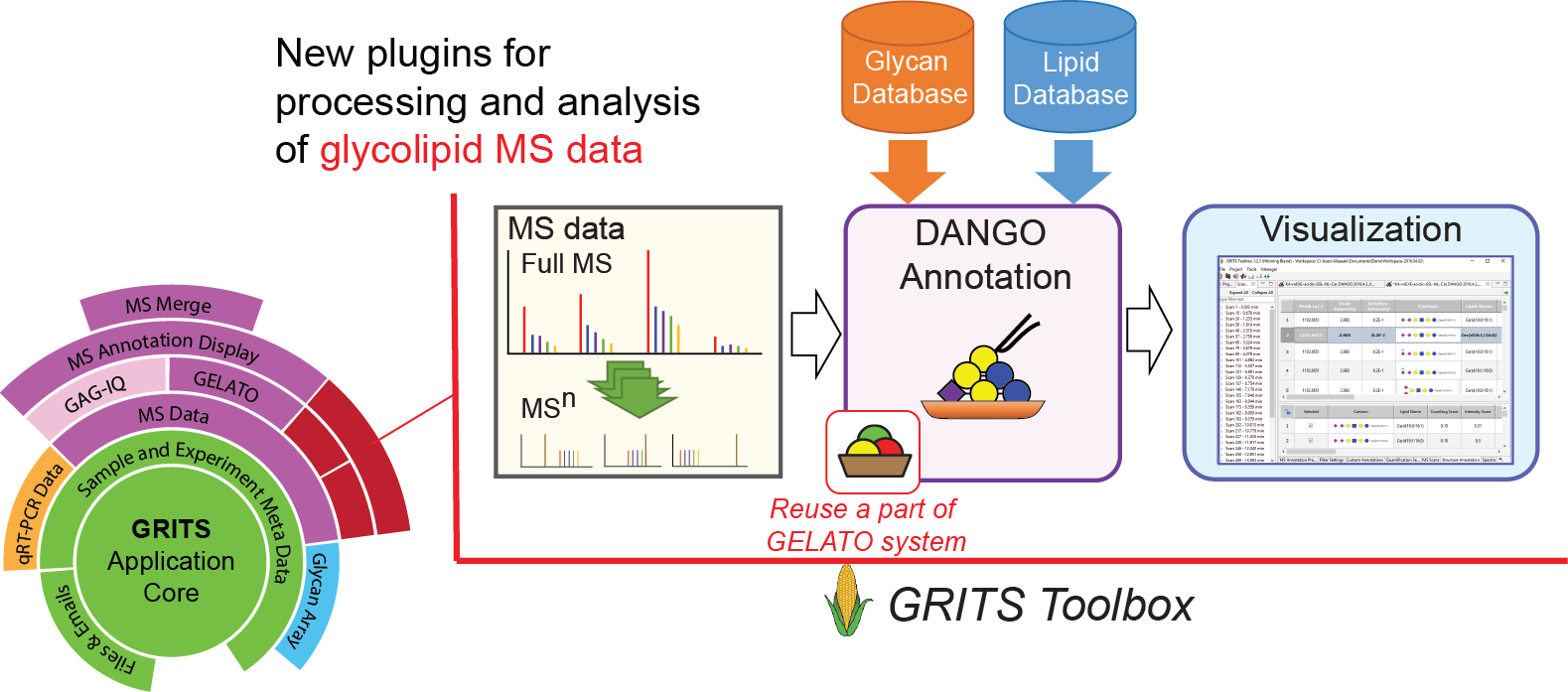
This protocol is complemented with our newly developed GSL data annotation software, calls DANGO (Data ANnotation system for GlycolipidOmics), which is being developed as an extension to the GRITS-Toolbox, an extendable software system for processing, interpreting and archiving glycomics MS data and associated metadata (http://www.grits-toolbox.org). The software annotates a dataset based on provided glycan and lipid databases and proposes candidate structures to the user, which can reduce the annotation time from weeks or months to hours.